-Search query
-Search result
Showing 1 - 50 of 155 items for (author: obr & m)
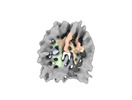
EMDB-19906: 
In situ nucleosome subtomogram average from Chlamydomonas reinhardtii
Method: subtomogram averaging / : Michael AK, Righetto RD, Obr M, van der Stappen P, Lamm L, Khavnekar S, Kotecha A, Engel BD
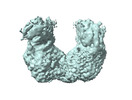
EMDB-17867: 
beta-Ureidopropionase tetramer
Method: single particle / : Cederfelt D, Dobritzsch D

PDB-8pt4: 
beta-Ureidopropionase tetramer
Method: single particle / : Cederfelt D, Dobritzsch D

EMDB-40477: 
KLHDC2 in complex with EloB and EloC
Method: single particle / : Digianantonio KM, Bekes M

PDB-8sh2: 
KLHDC2 in complex with EloB and EloC
Method: single particle / : Digianantonio KM, Bekes M

EMDB-29400: 
Acinetobacter baylyi LptB2FG bound to lipopolysaccharide and a macrocyclic peptide
Method: single particle / : Pahil KS, Gilman MSA, Kruse AC, Kahne D

EMDB-29401: 
Acinetobacter baylyi LptB2FG bound to lipopolysaccharide.
Method: single particle / : Pahil KS, Gilman MSA, Kruse AC, Kahne D

EMDB-29402: 
Acinetobacter baylyi LptB2FG bound to lipopolysaccharide and RG6006
Method: single particle / : Pahil KS, Gilman MSA, Kruse AC, Kahne D

EMDB-29403: 
Acinetobacter baylyi LptB2FG bound to lipopolysaccharide and a macrocyclic peptide
Method: single particle / : Pahil KS, Gilman MSA, Kruse AC, Kahne D

EMDB-29404: 
Acinetobacter baylyi LptB2FGC
Method: single particle / : Pahil KS, Gilman MSA, Kruse AC, Kahne D

EMDB-42206: 
Acinetobacter baylyi LptB2FG bound to Acinetobacter baylyi lipopolysaccharide
Method: single particle / : Pahil KS, Gilman MSA, Baidin V, Kruse AC, Kahne D

EMDB-42207: 
Acinetobacter baylyi LptB2FG bound to Acinetobacter baylyi lipopolysaccharide and a macrocyclic peptide
Method: single particle / : Pahil KS, Gilman MSA, Baidin V, Kruse AC, Kahne D

PDB-8frl: 
Acinetobacter baylyi LptB2FG bound to lipopolysaccharide and a macrocyclic peptide
Method: single particle / : Pahil KS, Gilman MSA, Kruse AC, Kahne D

PDB-8frm: 
Acinetobacter baylyi LptB2FG bound to lipopolysaccharide.
Method: single particle / : Pahil KS, Gilman MSA, Kruse AC, Kahne D

PDB-8frn: 
Acinetobacter baylyi LptB2FG bound to lipopolysaccharide and Zosurabalpin
Method: single particle / : Pahil KS, Gilman MSA, Kruse AC, Kahne D

PDB-8fro: 
Acinetobacter baylyi LptB2FG bound to lipopolysaccharide and a macrocyclic peptide
Method: single particle / : Pahil KS, Gilman MSA, Kruse AC, Kahne D

PDB-8frp: 
Acinetobacter baylyi LptB2FGC
Method: single particle / : Pahil KS, Gilman MSA, Kruse AC, Kahne D

PDB-8ufg: 
Acinetobacter baylyi LptB2FG bound to Acinetobacter baylyi lipopolysaccharide
Method: single particle / : Pahil KS, Gilman MSA, Baidin V, Kruse AC, Kahne D

PDB-8ufh: 
Acinetobacter baylyi LptB2FG bound to Acinetobacter baylyi lipopolysaccharide and a macrocyclic peptide
Method: single particle / : Pahil KS, Gilman MSA, Baidin V, Kruse AC, Kahne D

EMDB-16032: 
Structure of mouse heavy chain apoferritin solved by subtomogram averaging of 100 tilt series in Warp/M
Method: subtomogram averaging / : Obr M, Yang W, Karia D, Koh FA, Kotecha A

EMDB-15854: 
Structure of mouse heavy chain apoferritin solved by subtomogram averaging of 10 tilt series in Warp/M
Method: subtomogram averaging / : Obr M, Yang W, Karia D, Koh FA, Kotecha A

EMDB-18231: 
CryoDRGN Reconstruction of 80S Ribosome in Rotated State from S.cerevisiae Prepared using Cryo-plasmaFIB Milling
Method: subtomogram averaging / : Rangan R, Khavnekar S, Lerer A, Johnston J, Kelley R, Obr M, Kotecha A, Zhong ED

EMDB-18232: 
CryoDRGN Reconstruction of 80S Ribosome in Unrotated State from S.cerevisiae Prepared using Cryo-plasmaFIB Milling
Method: subtomogram averaging / : Rangan R, Khavnekar S, Lerer A, Johnston J, Kelley R, Obr M, Kotecha A, Zhong ED

EMDB-17929: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: pooled class
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17930: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -20 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17931: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -15 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17932: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -10 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17933: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -05 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17934: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 0 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17935: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 05 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17936: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 10 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17937: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 15 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17938: 
Cryo-electron tomogram of HTLV-1 MA126CANC tubes
Method: electron tomography / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17939: 
Cryo-electron tomogram of HTLV-1 MA126CANC tubes
Method: electron tomography / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17940: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 20 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17941: 
Cryo-electron tomogram containing HTLV-1 Gag-based VLPs
Method: electron tomography / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17942: 
Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

EMDB-17943: 
Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-CTD refinement
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

PDB-8pu6: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: pooled class
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

PDB-8pu7: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -20 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

PDB-8pu8: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -15 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

PDB-8pu9: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -10 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

PDB-8pua: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -05 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

PDB-8pub: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 0 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

PDB-8puc: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 05 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

PDB-8pud: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 10 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

PDB-8pue: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 15 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

PDB-8puf: 
Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 20 degrees
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

PDB-8pug: 
Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM

PDB-8puh: 
Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-CTD refinement
Method: subtomogram averaging / : Obr M, Percipalle M, Chernikova D, Yang H, Thader A, Pinke G, Porley D, Mansky LM, Dick RA, Schur FKM
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
